
Subunit Movements in Single Membrane-bound
H+-ATP Synthases from Chloroplasts during ATP Synthesis
Abstract
Subunit movements within the H+-ATP synthase from chloroplasts (CF0F1) are investigated during ATP synthesis. The γ-subunit (γCys-322) is covalently labeled with a fluorescence donor (ATTO532). A fluorescence acceptor (adenosine 5′-(β,γ-imino)triphosphate (AMPPNP)-ATTO665) is noncovalently bound to a noncatalytic site at one α-subunit. The labeled CF0F1 is integrated into liposomes, and a transmembrane pH difference is generated by an acid base transition. Single-pair fluorescence resonance energy transfer is measured in freely diffusing proteoliposomes with a confocal two-channel microscope. The fluorescence time traces reveal a repetitive three-step rotation of the γ-subunit relative to the α-subunit during ATP synthesis. Some traces show splitting into sublevels with fluctuations between the sublevels. During catalysis the central stalk interacts, with equal probability, with each αβ-pair. Without catalysis the central stalk interacts with only one specific αβ-pair, and no stepping between FRET levels is observed. Two inactive states of the enzyme are identified: one in the presence of AMPPNP and one in the presence of ADP.
Introduction
Membrane-bound H+-ATP synthases generate ATP from ADP and inorganic phosphate in bacteria, mitochondria, and chloroplasts. These enzymes use a rotational mechanism to couple proton transport through the membrane-integrated F0 part with ATP synthesis at the catalytic nucleotide-binding sites taking place in the hydrophilic F1 part (1,–4). Rotation of the γ-subunit labeled with actin filaments has been shown during ATP hydrolysis with single molecule techniques using immobilized F1 and F0F1 (5,–8). Rotation of the γ and ϵ-subunit during ATP synthesis has been shown with single-pair fluorescence resonance energy transfer (spFRET)2 (9) with membrane-integrated F0F1 (10,–12).
Single molecule investigations of the rotational mechanism have been carried out almost exclusively with bacterial F0F1 and its subcomplexes. Only a few results have been reported for eukaryotic H+-ATP synthases. Rotation of the γ-subunit in immobilized CF1 during ATP hydrolysis was shown spectroscopically with molecule ensembles (13), and it was visualized with single molecule techniques using labeled actin filaments (14). The redox regulation of rotation of the γ-subunit has been investigated with a chimeric F1 part constructed from bacterial α3β3 and the γ-subunit from chloroplasts (15). The physiological role of H+-ATP synthases in vivo is synthesis of ATP; however, subunit movements in CF0F1 during ATP synthesis have not been reported yet.
In this work we investigate subunit rotation in membrane-integrated CF0F1 in single enzymes using spFRET. Because cysteine mutants are not easily available, the enzyme was labeled with a fluorescence donor and an acceptor in the following way. An endogenous cysteine of the γ-subunit of CF0F1 was labeled selectively with the fluorescence donor (ATTO532). AMPPNP labeled at the ribose moiety with a fluorescence acceptor (ATTO655) was bound at a noncatalytic nucleotide-binding site on the α-subunit. The nucleotide-binding sites in CF1 have been extensively investigated (16). Labeling procedures have been reported for CF0F1 that allow a selective occupation of each catalytic and noncatalytic nucleotide-binding site (17,–19). The procedure reported for binding of ATP to noncatalytic site 4 has been adapted in this work for binding of AMPPNP-ATTO655. The donor- and acceptor-labeled CF0F1 was reconstituted into liposomes, and the movement of the γ-subunit relative to the α-subunit was measured with spFRET in freely diffusing proteoliposomes during proton transport-coupled ATP synthesis in a confocal microscope. These data show for the first time subunit rotation in eukaryotic H+-ATP synthases during ATP synthesis.
EXPERIMENTAL PROCEDURES
Isolation and Labeling of CF0F1
CF0F1 was prepared as described (20) and concentrated by ammonium sulfate precipitation. The pellet was resuspended in buffer A (20 mm Tricine-NaOH, pH 8, 20 mm succinic acid, 0.6 mm KOH, 0.1% (w/v) n-dodecyl-β-maltoside). For labeling of the γ-subunit, 8 μm CF0F1 was incubated with 4 μm ATTO532-maleimide (supplied by ATTO-TEC GmbH, Siegen, Germany) for 10 min in buffer A on ice. After the reaction, the free fluorophore was removed by passage through a Sephadex G-50 centrifugation column equilibrated with buffer A. The labeling efficiency was determined spectroscopically using the following absorption coefficients: ϵ532 = 115,000 m−1 cm−1, ϵ280 = 12650 m−1 cm−1 for ATTO532-maleimide (data from ATTO-TEC), and ϵ280 = 228,000 m−1 cm−1 for CF0F1 (calculated according to Ref. 21). The selectivity was checked by SDS gel electrophoresis. For selective labeling of the first noncatalytic site with AMPPNP-ATTO655 (ATTO-TEC), a modified procedure of Possmayer et al. (17) was used. CF0F1-γ-ATTO532 was integrated into liposomes (22) with a final enzyme concentration of 100 nm. These proteoliposomes were incubated with BioBeads (Bio-Rad) and 2 mm EDTA for 24 h at room temperature. The BioBeads were then removed, and AMPPNP-ATTO655 was added in a molar excess of 1:100 and incubated for 1 h at room temperature. Free AMPPNP-ATTO655 was removed by passage through a Sephadex G-50 centrifugation column equilibrated with buffer A. Then 2 mm EDTA and 10 μm ADP were added and incubated for 1 h at room temperature, and the free nucleotides were removed by a centrifugation column (see above). The labeling efficiency was measured by the fluorescence intensity of the bound AMPPNP-ATTO655. The ATP synthesis activity was measured at 23 °C (22, 23).
Single-pair Fluorescence Measurements
Single-pair fluorescence measurements were taken at room temperature using a two-channel confocal microscope set-up (24). The intensity of the laser beam (Nd:YAG, 532 nm; Coherent) was attenuated to 100 μW, and the concentration of the labeled CF0F1 liposomes was adjusted to 100 pm. Fluorescence was recorded with a multichannel scaler PC-card (PMS 300; Becker & Hickel) using a 1-ms binning time.
The measurements under noncatalytic conditions were carried out in a buffer containing 20 mm Tricine-NaOH, pH 8.0, 20 mm succinic acid, 2.5 mm MgCl2, 50 mm NaCl, 0.6 mm KCl, 4% (v/v) glycerol, and 1 mm AMPPNP. Measurements during ATP synthesis (15 or 100 μm ADP) were performed as described by Zimmermann et al. (11). Measurements during ATP hydrolysis were carried out after activation of the enzyme by ΔpH. Proteoliposomes (10 nm CF0F1) were incubated in buffer I (20 mm succinic acid-NaOH, pH 4.7, 5 mm NaH2PO4, 0.6 mm KOH, 2.5 mm MgCl2, and 10 μm valinomycin) for 3 min and this solution (10 μl) was mixed with 70 μl of buffer II (200 mm Tricine-NaOH, pH 8.8, 5 mm NaH2PO4, 160 mm KOH, 2.5 mm MgCl2, and 1 mm ATP) on a microscope slide. After 10 s the transmembrane pH difference was abolished by the addition of 10 μl of 30 mm NH4Cl, and the measurement was started. Data acquisition was carried out for 10 min.
FRET Data Analysis
Background count rates (usually between 0.5 and 2 kHz) obtained from measurements with pure buffer solution were subtracted. Additionally, cross-talk (donor signal leaking into the acceptor channel) and differences in the detection efficiencies of the donor and acceptor channel were taken into account to obtain the corrected fluorescence intensities of donor (FD) and acceptor (FA). The photon bursts had durations of 20 ms or longer and a minimum overall count rate of 400 photons. For each photon burst the FRET efficiency was calculated in 1-ms steps according to EFRET = FA/(FA + γFD) with a correction factor of γ = ηAΦA/ηDΦD, where ηD and ηA are the overall detection efficiencies of the donor and acceptor channel, respectively, and ΦD and ΦA are the fluorescence quantum yields of the enzyme-bound donor and the free acceptor, respectively. The detection efficiencies ηD = 0.37 and ηA = 0.37 were calculated by comparing the corrected emission spectrum of donor and acceptor with the transmission efficiencies of the filters and the spectral sensitivities of the detectors. The quantum yield of ATTO532 was determined to 0.43 ± 0.05 in reference to 5-carboxytetramethylrhodamine (25). Because of the low degree of labeling used in this work, the concentration of the enzyme-bound acceptor was below the detection level for quantum yield determination. Therefore, the quantum yield of the free acceptor was used for further calculations ΦA = 0.3 (specification of the manufacturer; ATTO-TEC). These data give a correction factor of γ = 0.70. In photon bursts with fluctuating FRET efficiencies, a constant FRET efficiency with a duration of t ≤ 5 ms was considered as a separate FRET level. The FRET levels for each condition (ATP synthesis, ATP hydrolysis, and AMPPNP binding) are presented in histograms.
Calculation of the Förster Distance between ATTO532 and ATTO655
The distance between donor and acceptor was calculated according to rDA = R0[(1/EFRET) − 1]1/6. In addition to the FRET efficiency, the Förster distance R0 was needed to calculate rDA. R0 was calculated according to R0 = 0.211[J(λ)·MD·κ2·λ4]1/6, where J(λ) is the overlap integral (26). The fluorescence excitation and emission spectra of ATTO532 and ATTO655 are shown in supplemental Fig. S2. The overlap integral describes the overlap of the emission spectrum of the enzyme-bound donor and the absorption spectrum of the acceptor (see orange area in supplemental Fig. S2). Evaluation of these data resulted in J(λ) = 2.8·10−13 m−1 cm3. MD = 0.43 is the quantum yield of the donor, and κ2 is the orientation factor between the emission transition dipole of the donor and the absorption transition dipole of the acceptor. Free rotation of the fluorophores was assumed, i.e. κ2 = 2/3, and with these data R0 = 53.1 Å was calculated.
RESULTS
Labeling and Activity of CF0F1
The γ-subunit of CF0F1 from spinach has four endogenous cysteines. The cysteines γCys-199 and γCys-205 form a disulfide bridge under oxidizing conditions, and γCys-89 is accessible only when the membrane is energized (27). The penultimate amino acid of the γ-subunit (γCys-322) has been labeled with ATTO532-maleimide, which binds under physiological conditions to cysteines. The SDS gel of the labeled CF0F1 and the fluorogram show the selective labeling of the γ-subunit without significant labeling of cysteines of the other subunits (supplemental Fig. S1). The labeled γ-subunit was extracted from the SDS gel, and mass spectrometric amino acid sequencing revealed that ATTO532 was bound to γCys-322. The degree of labeling was calculated as 20% from the absorption spectrum of the labeled CF0F1 as described under “Experimental Procedures.”
To obtain a double-labeled enzyme, AMPPNP-ATTO655, a nonhydrolyzable fluorescent ATP analogue was used. CF0F1 contains one noncatalytic nucleotide-binding site on each α-subunit. The binding properties of the three noncatalytic sites have been investigated, and methods for their selective occupation have been reported (17). Noncatalytic nucleotide-binding site 4 is the site with the highest binding affinity for ATP, and during catalytic turnover it does not exchange the bound ATP. Nucleotide-binding sites on CF1 have been characterized earlier, and the binding site with the highest affinity for ATP has been called site 2 (16, 28, 29); this site is identical with site 4 of CF0F1 (17). Therefore, this site was selected for noncovalent binding of AMPPNP-ATTO655. Exchange of the bound ATP against AMPPNP-ATTO655 was carried out as follows. CF0F1 was first labeled at γCys-322 with ATTO532, as described above, and then reconstituted into liposomes. Incubation of these proteoliposomes with EDTA and BioBeads for 24 h at room temperature partly removes the endogenous ATP from site 4. Then AMPPNP-ATTO655 was added, and after 1 h of incubation, free nucleotides were removed by passage through a centrifugation column. The labeling efficiency (30%) was estimated by comparing the ATTO655 fluorescence of the proteoliposomes with that of free AMPPNP-ATTO655, assuming the same quantum efficiency of the bound and free nucleotide.
The effect of the labeling procedures on enzyme activity was determined by measuring the rate of ATP synthesis. The labeled CF0F1 and, for comparison, unlabeled CF0F1 were reconstituted into liposomes. The proteoliposomes were energized by an acid base transition, and the initial rates of ATP synthesis were measured. The results are presented in Table 1. The rate obtained with CF0F1 was 140 s−1. When the labeling procedure for the γ-subunit was carried out without ATTO532, the rate decreased to 115 s−1. When ATTO532 was included, and the enzyme was labeled, the same activity was obtained independent of the degree of labeling. The procedure for occupation of noncatalytic site 4 (without AMPPNP-ATTO655) decreased the rate to 55 s−1. In the presence of AMPPNP-ATTO655, a similar activity (65 s−1) was observed. Therefore, in both cases the labeling procedure and not the presence of the label decreased the enzyme activity.
Subunit Movements during Catalysis
The spFRET measurements of the donor- and acceptor-labeled CF0F1 in liposomes were performed with a two-channel confocal microscope. The donor, ATTO532, was excited at 532 nm, and the fluorescence intensities of both donor and acceptor were measured simultaneously. The emission of the donor (FD) was collected in the wavelength range from 545 to 610 nm, the acceptor fluorescence (FA) was collected above 660 nm. When a liposome with a donor- and acceptor-labeled CF0F1 diffused through the confocal detection volume, photon bursts with fluorescence intensities up to 200 kHz (200 photons/ms) were observed. Because of the three-dimensional Gaussian profile of the focused laser, the trajectory of one proteoliposome through the confocal volume resulted in fluctuations of fluorescence intensities of both the donor and the acceptor (Fig. 1). However, the FRET efficiency, i.e. the ratio of their fluorescence intensities: IEFRET = FA/(FA + γFD), does not depend on the localization of the proteoliposome. Instead, it depends on the distance between donor and acceptor fluorophore, and its changes reflect relative subunit movements in the enzyme.
To observe ATP synthesis, the proteoliposomes are energized by an acid base transition. The mixing chamber is inserted into the confocal microscope, and photon bursts are observed up to 1 min after mixing. During ATP synthesis, in the presence of 100 μm ADP, ~20% of the bursts show rapid changes between the different FRET levels. Two examples of such bursts are depicted in Fig. 1 (A and B), showing the FRET level sequence L → M → H from low (L) to medium (M) and to high (H) FRET efficiency. This sequence is observed in 30% of the bursts with FRET transition. Fig. 3A depicts a histogram of bursts with changing FRET levels. All three levels occur with the same probability (33%). The three peaks are fitted with Gaussian distributions resulting in maxima at EFRET(L) = 0.26 ± 0.01, EFRET(M) = 0.57 ± 0.01, and EFRET(H) = 0.84 ± 0.01. The mean donor acceptor distances are calculated from the maxima (see “Experimental Procedures”), resulting in 6.3 nm (L), 5.1 nm (M), and 4.0 nm (H).
In addition to bursts with three FRET levels, bursts with a number of sublevels were observed (54% of the bursts with FRET transition). An example is shown in Fig. 1C. These bursts also show the sequence L → M → H; however, each level is split into two sublevels, and forward and backward steps between the sublevels are detected (e.g. L1 → L2 → L1 at 160 ms and M2 → M1 → M2 at 320 ms in Fig. 1C). Furthermore, rather than a stepwise transition, some traces show an almost continuous variation in FRET efficiency (Fig. 1D).
Similar measurements have been carried out during ATP synthesis in the presence of 15 μm ADP. Also in this case bursts with three changing FRET levels are observed, resulting in a distribution similar to that shown in Fig. 3A. The maxima of the three Gaussian distributions are found at the same FRET efficiencies and distances as in the presence of 100 μm ADP. The statistics of all photon bursts are shown in supplemental Tables S1 and S2.
Some experiments have been carried out during ATP hydrolysis in the presence of 1 mm ATP. In addition to bursts with constant FRET efficiency, bursts with changing FRET levels are observed. Their sequence is opposite to that during ATP synthesis, as observed with EF0F1 (10, 11).
The time course of FRET efficiency directly reflects the kinetics of subunit movements within CF0F1. A constant FRET efficiency indicates the docking of the γ-subunit to one specific αβ-pair (“dwell time”). Presumably, the dwell time is the time for synthesis of one ATP at one catalytic site. Switching to the next FRET state occurs faster than the time resolution of the measurements (1 ms). Thus, the movement of the γ-subunit in CF0F1 is a three-step rotation similar to that observed with EF0F1 (10). The arithmetic mean value of all dwell times was calculated, resulting in 29 ms in the presence of 100 μm ADP and 56 ms in the presence of 15 μm ADP. The statistics of the dwell times are shown in supplemental Fig. S3. These dwell times correspond to rates of 34 and 18 s−1. In comparison with the ensemble rate of the donor- and acceptor-labeled enzyme (24 s−1 in the presence of 100 μm ADP), the turnover obtained from single molecule measurements is somewhat higher (Table 1). This is expected because inactive enzymes are present in ensemble measurements, whereas in single molecule experiments only the active enzymes are selected.
Some bursts with FRET transitions show the sequence H → M → L (17% in the presence of 100 μm ADP, 5% in the presence of 15 μm ADP). These “wrong sequences” (see supplemental Table S1) are due to the combination of short turnover times of ATP synthesis and the fact that only FRET levels with durations longer than 5 ms are considered to be separate levels. Assuming an exponential distribution of the dwell times, the probability of missing a FRET level with a dwell time shorter than 5 ms can be calculated using N/N0 = (1 − exp(−t/τ)). Using t = 4 ms, we obtain with τ = 29 ms (dwell time in the presence of 100 μm ADP) a fraction of 13%; with τ = 56 ms (dwell time in the presence of 15 μm ADP), we obtain a fraction of 7%. The loss of one level in the correct sequence leads automatically to the opposite sequence. Because the fraction of levels with a dwell time shorter than 5 ms increases with the rate, the fraction of wrong sequences increases with the rate of ATP synthesis as observed. We conclude from this consideration that nearly all bursts during ATP synthesis show the same FRET level sequence (10).
Relative Subunit Positions without Catalysis
During ATP synthesis, i.e. within 1 min after mixing, ~80% of the bursts show a constant FRET efficiency. Obviously, these enzymes do not perform catalysis. There might be two reasons for this behavior. Either the enzymes are inactive, or they are still active but the transmembrane pH difference has declined. In the latter case the rate of ATP synthesis is low, leading to a high dwell time so that the proteoliposomes diffuse through the confocal volume without a FRET transition. After approximately 1 min the transmembrane pH difference has declined so that ATP synthesis is not possible, and nearly all bursts observed later than 1 min after mixing show a constant FRET efficiency. In the following text these conditions are referred to as “after ATP synthesis.” The observed signals are depicted in Fig. 2 (A–D). The histogram showing bursts after ATP synthesis is presented in Fig. 3 B. Four different FRET levels are found: low (L, 34%), medium (M, 7%), high (H, 12%), and most frequently (HADP*, 42%). The Gaussian distributions show maxima at EFRET(L) = 0.24 ± 0.02, EFRET(M) = 0.55 ± 0.02, EFRET(H) = 0.84 ± 0.02, and EFRET(HADP*) = 0.95 ± 0.08. The donor acceptor distances are 6.4 nm (L), 5.1 nm (M), 4.0 nm (H), and 3.3 nm (HADP*), respectively. Comparison of these distances with those obtained from bursts with changing FRET levels indicates that the L, M, and H states during and after ATP synthesis are equal. Presumably, these constant levels represent enzymes that either do not rotate during the burst or have finished catalysis. They are obviously in the same conformation as during ATP synthesis, i.e. they have the same three conformations as during catalysis, although they do not show FRET transitions.
The most frequent FRET level, HADP*, is a new conformation. In a few cases a transition from the level H to HADP* is found. We did not detect photon bursts where a transition from HADP* into another state was observed. Obviously, this conformation is very stable and is only observed when no catalysis takes place. We assume that it represents an inactive state of the enzyme containing a tightly bound ADP.
AMPPNP is an ATP analogue that binds to CF0F1 but is not hydrolyzed. The spFRET measurements in the presence of AMPPNP show photon bursts with constant levels. Bursts with low (L) and medium (M) FRET efficiency are very rare; nearly all bursts show high (H) FRET efficiency (Fig. 2E). A histogram of the number of FRET levels as a function of FRET efficiency is depicted in Fig. 3C. The highest FRET level (HAMPPNP*) is observed most frequently (74%). This state of the enzyme is energetically favored and represents a stable, inactive conformation. Fitting the data to a Gaussian distribution gives the most probable FRET efficiency, EFRET = 0.98 ± 0.02, resulting in a donor acceptor distance of 2.8 nm. The number of bursts with other FRET levels is too low for further evaluation (Fig. 3C). These data provide evidence for two different inactive states of the enzyme, which are characterized by their different donor acceptor distances: one in the presence of ADP (HADP*) and one in the presence of AMPPNP (HAMPPNP*).
DISCUSSION
Catalytic and Noncatalytic Conformations
The movement of the γ-subunit relative to the α-subunit during ATP synthesis within membrane-integrated CF0F1 was investigated with single-pair FRET. The donor fluorophore ATTO532 was covalently bound at the endogenous γCys-322. The acceptor fluorophore AMPPNP-ATTO655 was noncovalently bound to the noncatalytic nucleotide-binding site 4. Site 4 has been chosen for binding of AMPPNP-ATTO655 because it exchanges the bound ATP very slowly (17,–19).
The spFRET measurements revealed: 1) When a transmembrane ΔpH is generated by an acid base transition, ATP synthesis is observed for maximally 1 min after mixing because ΔpH decreases continuously. During ATP synthesis photon bursts with FRET transitions are observed. Three equally populated FRET states are observed with mean distances of 6.3 nm (L), 5.1 nm (M), and 4.0 nm (H). These distances are characteristic for enzymes in the active state during catalysis. Similar results are found in the presence of 15 and 100 μm ADP. The average dwell time, however, changes from 29 to 56 ms in accordance with the dependence of the turnover on ADP concentration. 2) Analysis of photon bursts after ATP synthesis reveals four constant FRET efficiencies with the population: 34% L, 7% M, 12% H, and 42% HADP*. The maxima of the Gaussian distributions correspond to the distances 6.4 nm (L), 5.1 nm (M), 4.0 nm (H), and 3.3 nm (HADP*). The L, M, and H states have, within error limits, the same distances during and after ATP synthesis. These enzymes do not carry out catalysis, but they have the same conformations as the active enzymes. The situation is different for the HADP* state. The distance is significantly lower than during catalysis (3.3 nm), and this state is accumulated with time after catalysis. Presumably, this represents an inactive state. 3) Photon bursts in the presence of AMPPNP display no FRET transitions. Nearly all of the enzymes are found in the same FRET level, i.e. they are in one specific conformational state (HAMPPNP*), with a distance of 2.8 nm between γCys-322 and the noncatalytic site. This represents an inactive state.
Asymmetry of the Enzyme
According to the rotational binding change mechanism, all of the catalytic nucleotide-binding sites on the three αβ-pairs of F1 are intrinsically equivalent, and only the different interactions of each αβ-pair with the centrally located γ-subunit change their properties. Therefore, at any given time each pair acquires a different conformational state.
In the holoenzyme the situation is different: one αβ-pair is connected with the peripheral stalk (subunits I, II, δ), and this interaction might change the conformation of the tagged αβ-pair. If tagging changes the conformation of one αβ-pair, the tagged pair might interact with the γ-subunit in a stronger, weaker, or unchanged manner compared with the nontagged pairs. A stronger interaction would imply that the γ-subunit interacts preferentially with the tagged αβ-pair, i.e. there is one stable orientation of the γ-subunit to the peripheral stalk. A weaker interaction implies that the interaction with the two untagged αβ-pairs is more stable. If the interaction is unchanged, three stable positions of the γ-subunit relative to the peripheral stalk are expected.
The structure of CF0F1 shows the three different positions of γCys-322 relative to noncatalytic site 4 on the α-subunit. The spFRET data give information about the stability of the interaction of the γ-subunit with the different αβ-pairs. During ATP synthesis the three different FRET levels are populated with equal probability (Fig. 3A), i.e. all αβ-pairs interact similarly with the γ-subunit as proposed by the binding change theory (3). After ATP synthesis, when ΔpH has decreased below the threshold necessary for ATP synthesis, FRET level M is weakly populated, whereas L and H are more populated, i.e. L and H have a slightly stronger interaction with the γ-subunit (Fig. 3B). The FRET level HADP* is found most frequently, and it represents the conformation of a stable inactive state. In the presence of AMPPNP, nearly all of the enzymes are found in the state HAMPPNP* (Fig. 3C). This implies that the interaction of the γ-subunit with one αβ-pair is much stronger than that with the other αβ-pairs. Because no ATP synthesis can be observed under these conditions, it represents an inactive state of the enzyme.
Mellwig and Böttcher (30) used cryo-electron microscopy data to calculate a three-dimensional structure of CF0F1 in presence of AMPPNP with a resolution of 2 nm and used this as a template for fitting the homology models of subunits α, β, and γ. It was found that the optimal fit of the central stalk can be obtained only in one position of the γ-subunit relative to the peripheral stalk. In the inactive state of CF0F1, the γ-subunit has only one stable resting position relative to the peripheral stalk. This is in agreement with the findings from this study that only one FRET state is found in the presence of AMPPNP. Obviously, this position is energetically favored. It is concluded from the optimal fit that subunit αT is tagged by the peripheral stalk (30).
The distances between γCys-322 to the noncatalytic sites on the α-subunits were calculated from the homology model. The shortest distance was 4 nm in accordance with the FRET data obtained during ATP synthesis. In addition, two inactive states were found: one in the presence of ADP and one in the presence of AMPPNP. These were characterized by distances between γCys-322 and noncatalytic site 4 of 3.3 and 2.8 nm, respectively. The distance between the two fluorophores is shorter in the inactive state than in the active state during catalysis. This is in accordance with earlier results, where a decrease of the molecular volume of the F1 part induced by nucleotide binding was observed by electron microscopy (31) and fluorescence correlation spectroscopy (32).
Flexibility of the γ-Subunit
Compared with the three-step rotation sequence of the γ-subunit found in membrane-integrated F0F1 from Escherichia coli, the FRET sequences observed with CF0F1 show two additional features. First, the levels are split frequently into two sublevels with forward and backward steps between the sublevels. Second, continuous changes from low to high FRET are observed in some cases. The splitting of the docking dwell of γ at one αβ-pair into two dwell times has been observed earlier during ATP hydrolysis (8), and they have been attributed to an “ATP waiting dwell” and a “catalytic dwell.” During ATP synthesis, an ADP waiting dwell, a proton waiting dwell, and a catalytic dwell might be expected. In some photon bursts continuous FRET changes are detected. We think that the docking and undocking of the γ-subunit near the catalytic sites is always a stepwise process as observed earlier in EF0F1 during ATP synthesis and ATP hydrolysis (10, 11). In contrast to EF0F1, in CF0F1 the fluorophore was attached at the C terminus of the γ-subunit. There is a long distance between the C terminus and the torque-generating sites. Presumably, the flexibility of the long α-helices leads to a dumping of the stepped movement, at least in some conformations of the enzyme, so that a quasi-continuous movement is observed. It has been shown that a large part of the γ-subunit can be removed without changing the torque generation (33). Additionally, the flexibility of the γ-subunit near the top of the enzyme is so large that it can be covalently linked to the α-subunit without impairing the function (34).
Activation
The activity of CF0F1 is controlled by a redox reaction and a ΔpH-dependent shift from an inactive into an active state (35). The redox reaction is the formation of a disulfide bond between γCys-199 and γCys-205 and its reduction (36). The ΔpH-dependent activation is connected with the release of ADP from a catalytic site (37).
In single molecule spectroscopy, the FRET states of active enzymes are identified by the occurrence of FRET transitions; by mean dwell times, which are in accordance with the turnover time of the enzyme; and by the equal population of the three FRET states. The inactive enzymes are identified by a FRET efficiency distinct from that of active enzymes, a constant FRET level without transitions, and the fact that all enzymes are found in a single FRET state. In some photon bursts a FRET transition from the active state (H) into the inactive state (HADP*) was observed. Therefore, the γ-subunit not only plays a central role in the rotational binding change mechanism, it is also involved in the activation/inactivation process of the enzyme. The spFRET data show that ΔpH-dependent activation is a conformational change in CF0F1, which results in an increase in the distance between γCys-322 and noncatalytic site 4. This is accompanied by a decrease in the binding strength between the γ-subunit and one αβ-pair, so that in the active state, the rotating γ-subunit tags all three αβ-pairs with equal probability. It is only in this active state that catalysis can take place as described by the binding change mechanism.
Distances from the Homology Model and FRET Measurements
To compare the distances calculated from the FRET data with structural data, the homology model of CF0F1 (30, 38) was used. Fig. 4 shows the F0 part and one αβ-pair from this homology model. The α-subunit tagged by the peripheral stalk is αT. In addition βE is depicted because βETyr-385 is part of the noncatalytic binding site on αT (1). The following assumptions are used for this comparison. The noncatalytic binding site 4 is located on αT. The axis of rotation (red vertical line in Fig. 4A) is defined by the center of the αβ-hexamer. The stator label is positioned at amino acid βEY385 (red sphere). To obtain an estimation of the localization of the rotor label, we use γCys-321 because this is the last resolved amino acid in the x-ray structure of the C-terminal part. Moreover we take into account the size of the label including the linker (0.7 nm). If we displace γCys-321 for 0.7 nm by a counter-clockwise rotation (viewed from CF0 to CF1) in connection with a subsequent rotation of two 120° steps, we yield three positions of γCys-321 (see green spheres in Fig. 4, B and C), whose distances to βTyr-385 are in a good agreement with the experimental data. The obtained distances are 6.1, 4.8, and 3.9 nm (see supplemental Table S2). However, this agreement should not be overestimated. First, the stator label is located at the noncatalytic nucleotide-binding site 4, but the exact position of the transition dipole moment of the acceptor fluorophore is not known. Second, the position of the rotor label (γCys-322) is not detected by x-ray crystallography (1), obviously because of the large flexibility of the C-terminal part of the γ-subunit. This flexibility was shown by covalently linking the C-terminal end of the γ-subunit to the α-subunit without blocking the function of the enzyme (34).
Nevertheless the data show a rotational movement of the subunit in 120° steps during ATP synthesis relative to the α-subunit with the FRET level sequence L → M → H → L. When viewed from CF0 to CF1, this corresponds to a clockwise movement of the γ-subunit during ATP synthesis and a counter-clockwise movement during ATP hydrolysis (Fig. 4C). This direction of intersubunit rotation during hydrolysis within the holoenzyme is in accordance with earlier observations at the CF1 part (14). We are aware of the fact that these considerations do not provide the absolute direction of rotation. Therefore, the absolute direction is based on earlier direct determinations during ATP hydrolysis on single CF1 parts (14).
Acknowledgments
We gratefully thank F. Drepper and W. Haehnel (Department of Biology II, University of Freiburg, Freiburg, Germany) for mass spectrometry of the labeled enzyme.
 The
on-line version of this article (available at http://www.jbc.org/)
contains supplemental
Tables S1 and S2 and Figs. S1–S3.
The
on-line version of this article (available at http://www.jbc.org/)
contains supplemental
Tables S1 and S2 and Figs. S1–S3.- spFRET
- single-pair fluorescence resonance energy transfer
- AMPPNP
- adenosine 5′-(β,γ-imino)triphosphate
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
Figures and Tables
TABLE 1
Rate of ATP synthesis of reconstituted CF0F1 at 23 °C, measured at different states during the preparation procedure
The measurements were carried out with 100 μm ADP present in buffers I and II. The numbers are the averages of three measurements with standard deviations. The last two lines show the catalytic rates obtained from average dwell times of single molecule experiments (supplemental Fig. S3).
| Enzyme | Rate |
|---|---|
| s−1 | |
| CF0F1 | 140 ± 15 |
| CF0F1 after treatment for γ labeling | 115 ± 5 |
| CF0F1 γCys-322−ATTO532, 16% labeling | 120 ± 10 |
| CF0F1 γCys-322−ATTO532, 48% labeling | 115 ± 8 |
| CF0F1 after treatment for occupation of site 4 | 55 ± 5 |
| CF0F1 after occupation of site 4 with AMPPNP-ATTO655 | 65 ± 10 |
| Donor- and acceptor-labeled CF0F1 | 24 ± 6 |
| Donor- and acceptor-labeled CF0F1 average dwell time (100 μm ADP) of 29 ms | 34 ± 4 |
| Donor- and acceptor-labeled CF0F1 average dwell time (15 μm ADP) of 56 ms | 18 ± 3 |
FIGURE 1.
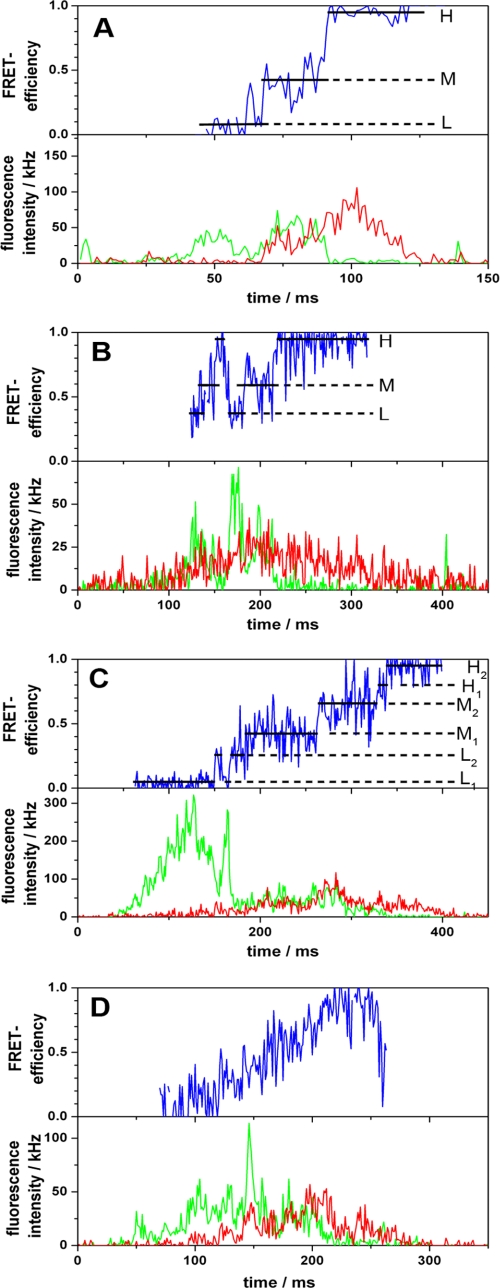
Photon bursts of FRET-labeled CF0F1 in liposomes during ATP synthesis. Corrected fluorescence intensity traces of the donor and acceptor are depicted in green and red, respectively. The FRET efficiencies (EFRET) calcu lated from these traces are shown as blue traces. For each FRET level, the arithmetic mean value is calculated (black lines). The FRET states are labeled L, M, and H. A and B, the three-step FRET level sequence is L → M → H → L. C, FRET level sequence with sublevels. D, continuous FRET changes.
FIGURE 2.
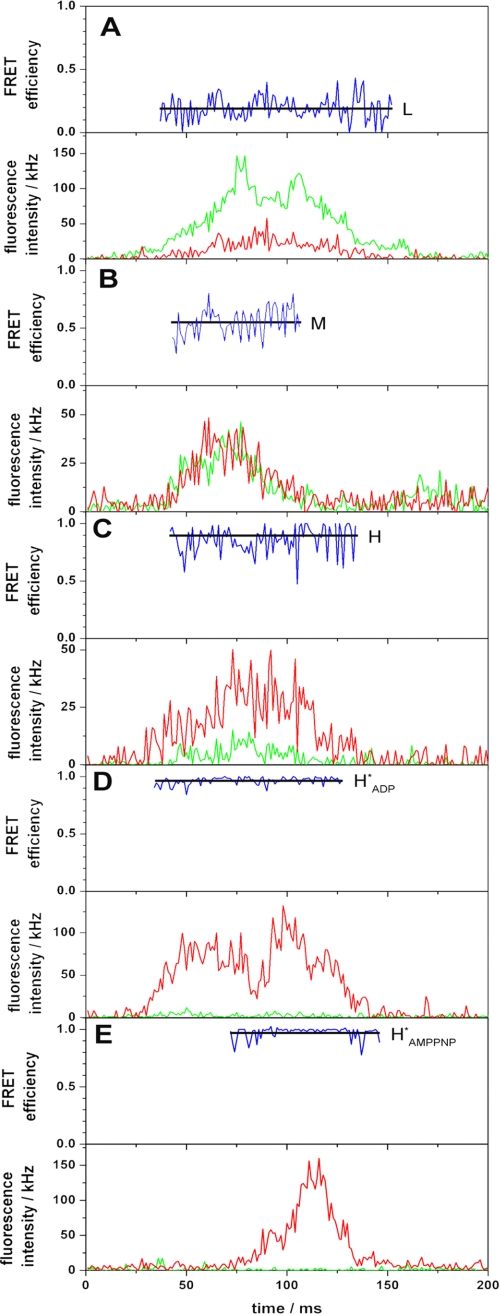
Photon bursts of FRET-labeled CF0F1 in liposomes in the presence of AMPPNP. Corrected fluorescence intensity traces of the donor (ATTO532) and acceptor (ATTO665) are depicted in green and red, respectively. The FRET efficiencies (EFRET) calculated from these traces are shown as blue traces. For each FRET level, the arithmetic mean value is calculated (black lines). The FRET states are labeled L (low), M (medium), H (high), HADP* (inactive state with bound ADP), and HAMPPNP* (inactive state with bound AMPPNP).
FIGURE 3.
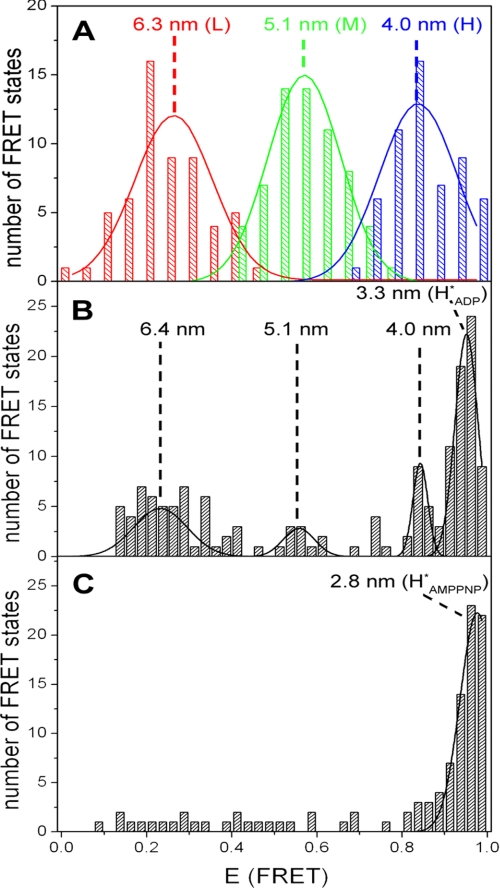
Histograms of the FRET efficiencies for CF0F1. The mean FRET efficiency is calculated for each observed FRET level (see black lines in Figs. 1 and and2),2), and the number of observations is plotted as a function of EFRET. The calculated donor acceptor distances are given for the maximum of each FRET level population. A, ATP synthesis with 100 μm ADP (62 photon bursts). The peaks are fitted by Gaussian distributions. B, after ATP synthesis in presence of 100 μm ADP (150 bursts). C, in the presence of 1 mm AMPPNP (data from 105 photon bursts).
FIGURE 4.
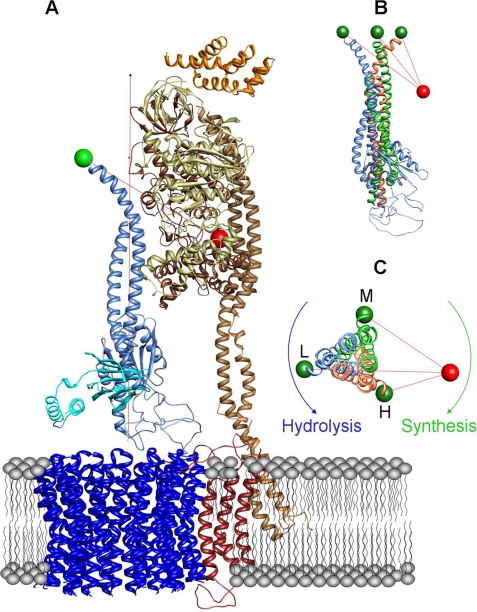
Visualization of the donor acceptor distances in the CF0F1 model. A, homology model of the CF0F1 holoenzyme (38). Only αT and βE are shown for clarity. The peripheral stalk (subunits I and II) is attached to the αT-subunit. The acceptor is located at Tyr-385 (red sphere) of the βE-subunit. The donor (green sphere) is located at Cys-321 of the γ-subunit (blue). The axis of rotation is indicated by a vertical red line. The γ-subunit is shown in the position with the largest donor acceptor distance (L). B, three positions of the γ-subunit in the same view as in A. A rotation of the γ-subunit in 120° steps leads from the original position (blue, same orientation as in A to the orange and green colored orientation of the γ-subunit. The distances between the three donor positions and the acceptor position are indicated by red lines. C, same model as in B but viewed from CF0 into the direction of CF1.