Electrophoresis
-
similar to SDS PAGE for proteins: (acrylamide--chemical crosslinks)
-
usually on a horizontal agarose gel--melts at high temperature; solid at
room temp
-
detect DNA using ethidium bromide/UV light
-
also detect using radioactivity (32P); fluorescence; chemiluminescence
|
-
nucleic acids are negatively charged, so move
- toward + electrode according
to their charge
-
proportional to MW--separate according to size
-
smaller NA move more quickly through gel than larger NA
|
Restriction enzyme--molecular scissors
-
endonucleases--does not require an end (exonucleases)
-
>100 restriction enzymes known
-
names come from organism:
-
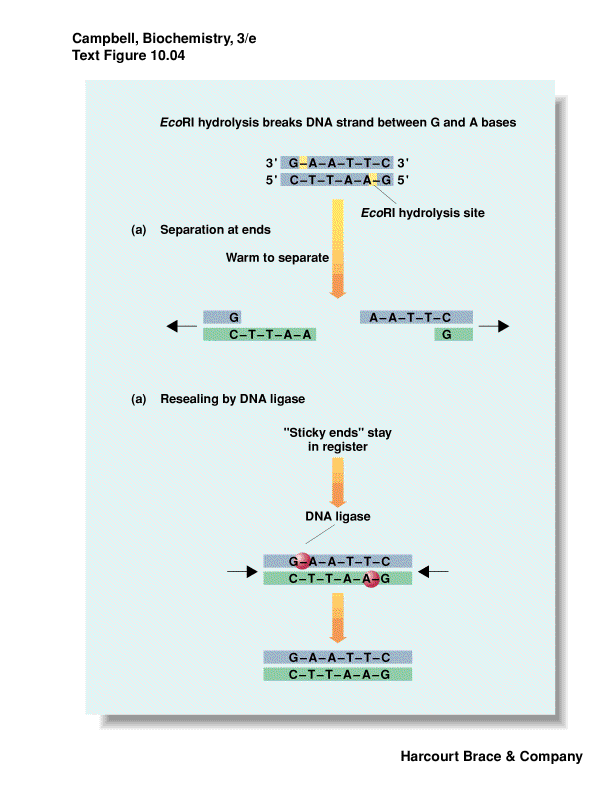
recognize a specific palindromic DNA sequence and cut the DNA
-
palindrome is the same forwards/backwards
-
some leave 3' overhang; 5' overhang or blunt ends
-
overhangs leave--"sticky ends"--even though DNA is cut, can have base-pairing
-
move DNA from one organism to another - "recombinant DNA"
-
put DNA together with DNA ligase
-
use synthetic DNA of desired sequence to "paste" on restriction site if
nature did not provide one
|
-
methylation protects DNA from restriction enzymes
-
mechanism for bacteria to protect itself from invading phage or other bacterial
DNA
 |
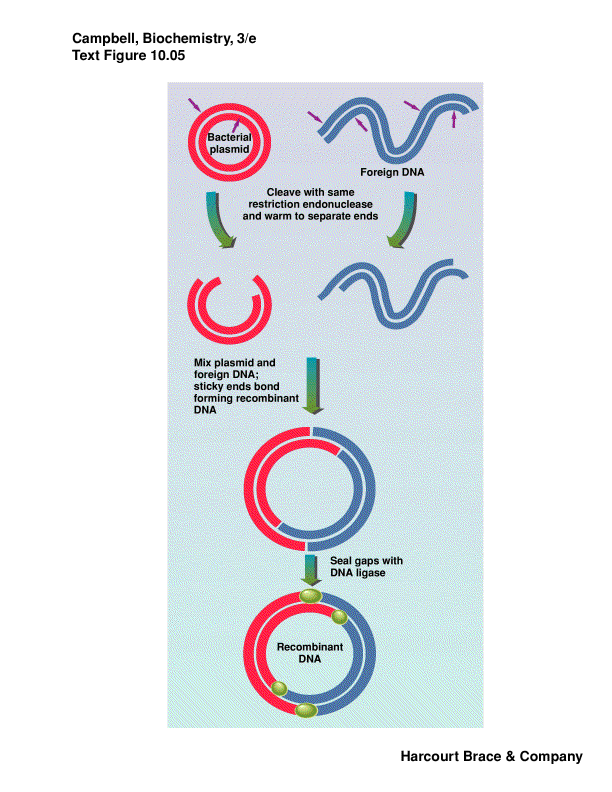
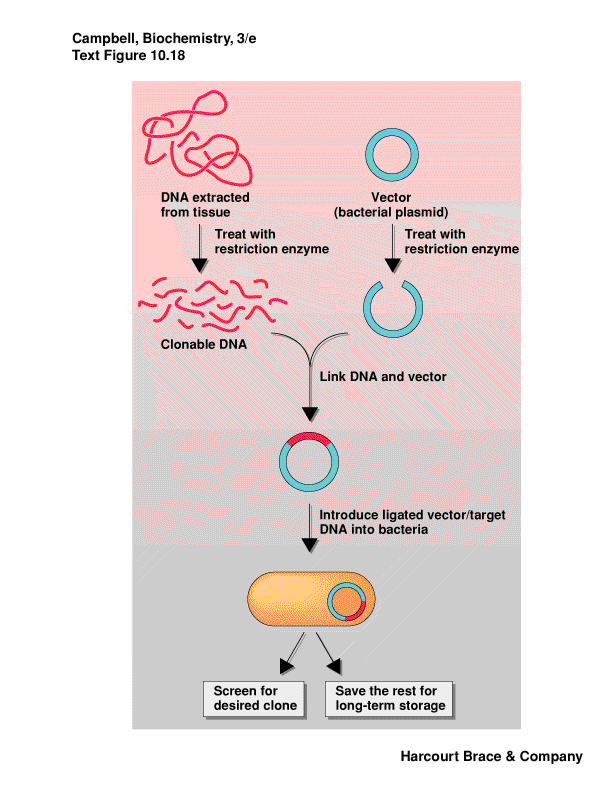
Plamids are cloning vectors
-
plasmids are closed circular DNA, with origin of replication--replicated
within bacteria to many copies
-
carries a resistance gene--ampicillin, tetracyclin, kanamycin
-
take DNA from one organism, cut with RE, isolate fragment desired from
a gel
-
cut a plasmid or phage DNA with same RE
-
put these two DNA fragments together via sticky ends, ligate them closed
-
we have recombinant DNA
-
this is transferred into bacterial cells by electroporation or chemical
competence
-
plate on media with antibiotic to kill bacteria that did not take up a
plasmid--no proof that your foreign DNA is there, only that the plasmid
is there
-
individual colonies contain a single plasmid
|
 |
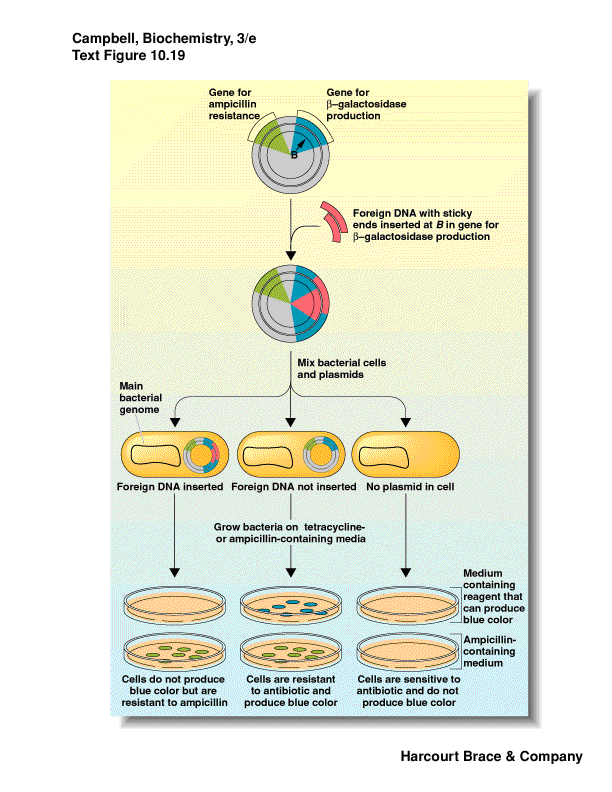
-
How do you know your foreign DNA was inserted?
-
one method: interrupt a gene that is a reporter - b-galactosidase (lacZ)
-
use a substrate for b-galactosidase that when
cleaved give a colored compound
-
do this on antibiotic media to select for plasmid
-
induce the gene with a lactose-analog
-
if the gene is intact get blue color--no foreign insert, just plasmid
-
if the gene has an insert (foreign DNA) then the reading frame is thrown
off and no b-galactosidase is produced--no color
|
 |
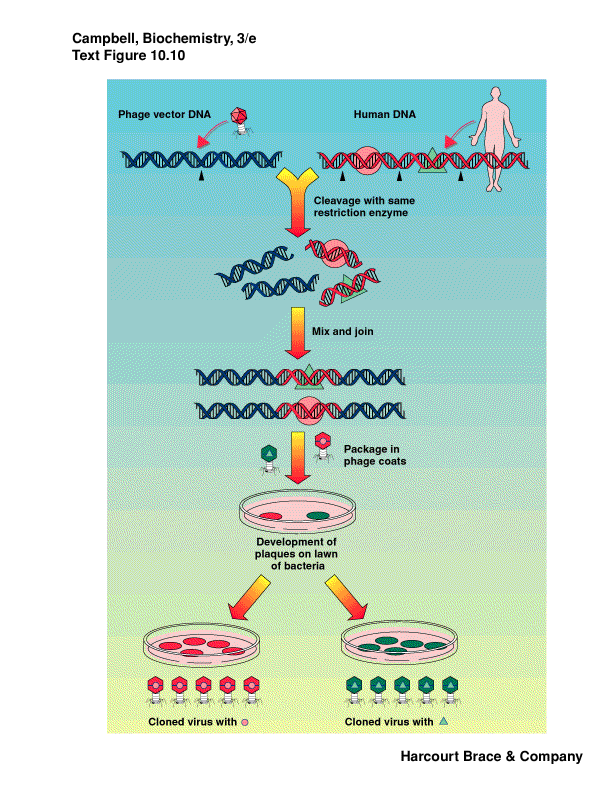
-
phage may be used as vectors
-
phage are bacterial viruses
-
accept larger pieces of DNA
-
same procedures of cut/paste
-
package phage DNA into virus particles
-
infect a lawn of bacteria
-
phage infection lyses bacteria, clear area "plaque"
-
each plaque represents a single phage
|
 |
DNA Libraries--what is the purpose of the library?
-
genomic DNA that has been fragmented by RE
-
will include introns from eukaryotes
-
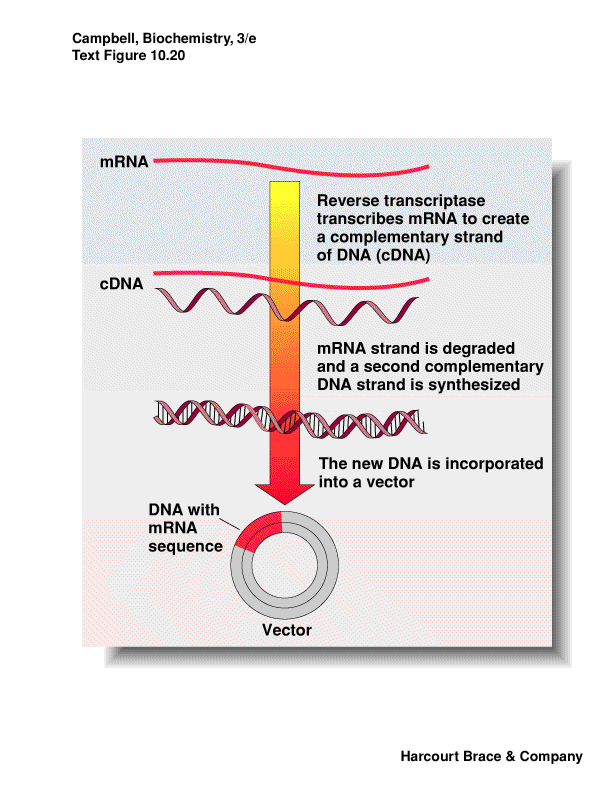
cDNA made from mRNA
-
for eukaryotes solve the problem of introns
-
similar methods of cut/paste into desired vector: plasmid vs. phage
-
phage easier to screen larger numbers; larger inserts; pain to isolate
and purify DNA from
-
plasmids are easier to work with once identified
|
  |
Screening of colonies/plaques
-
how do you find the gene you want?
-
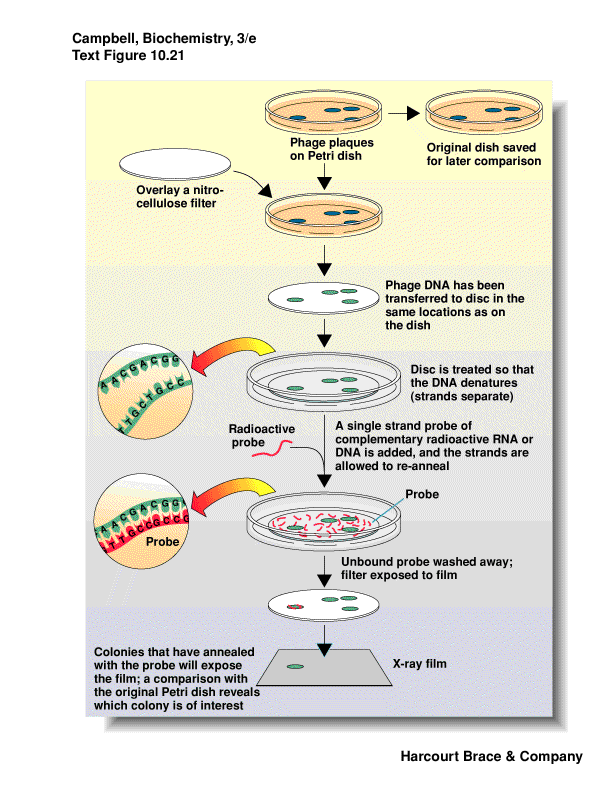
need a probe
-
a similar gene from another organism if the genes are similar enough
-
synthetic DNA based upon a protein sequence
-
need to take into account the degeneracy of genetic code
-
label the probe with radioactivity or a molecule that can be detected (e.g.
biotin)
-
transfer plaques or phage to a solid support (nitrocelluose or nylon)
-
denature DNA into single strands
-
probe will anneal to DNA it is complimentary to
-
detect which plaque or colony contains desired DNA
-
retrieve the plaque or colony from original plate
|
 |
Applications: What can you do with a gene once you have it?
-
ask bacteria to make the protein for you
-
design in the appropriate promoter, Shine-Delgarno sequences to get transcription/translation
-
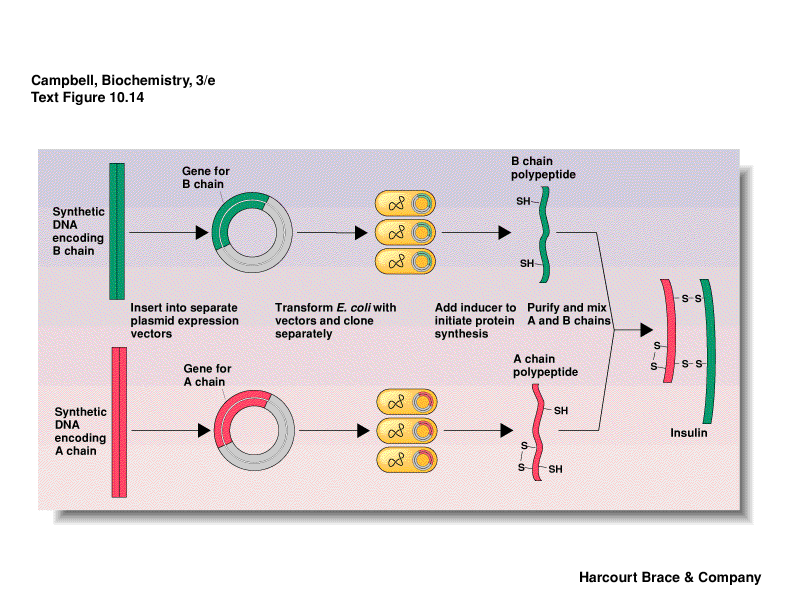
insulin is made by bacteria
-
make chains separately; join by disulfide bonds
-
human growth hormone; tissue plasminogen activator/enterokinase (dissolve
blood clots); erythropoietin (stimulates RBC production); many others
|
 |
Transgenic Organisms
-
put the gene back into an organism
-
put the gene into the germ line; inject DNA into the nucleus of fertilized
eggs; recombination
-
search out the gene in the population of offspring
-
this gene is now carried in the germ line and passed to succeeding generations
-
use similar techniques to knockout a gene and ask what effect it has on
the organism
|
Agricultural
Transgenic Plants
-
take a gene
from one organism and put in a different type
-
artic fish anti-freeze protein and put into frost-sensitive plants --protects
from freezing
-
insect toxin, Bt, express in plant to prevent insect damage: corn,
cotton
-
insert resistance to herbicides--soybeans
-
improve nutritional quality--"golden" rice--vitamin A in rice
-
prevent viral diseases
-
learn more about agricultural
biotechnology
|
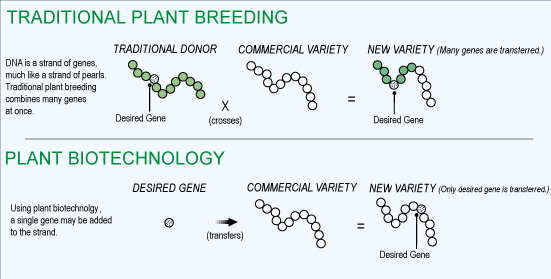
Is this bad, are these Frankenfoods?
-
plants and animals have been bred for thousands of years for desirable traits
-
trial and error to observe/produce the trait wanted
-
biotech simply shortens the time needed to introduce the trait
|
 |
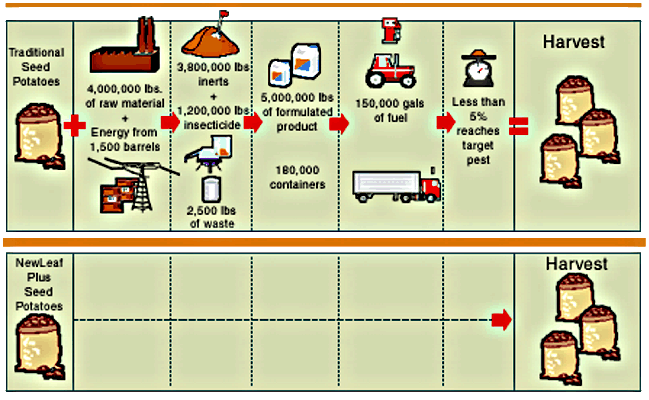
-
Compare enegy needed to grow potatoes conventionally with pesticides vs
with Bt
-
do we need to be cautious--yes--products should be well tested before released
-
there are many benefits to be gained
-
better nutrition
-
fewer chemical pesticides
-
pharmaceuticals--edible vaccines
-
be informed and make a rational choice
|
 |
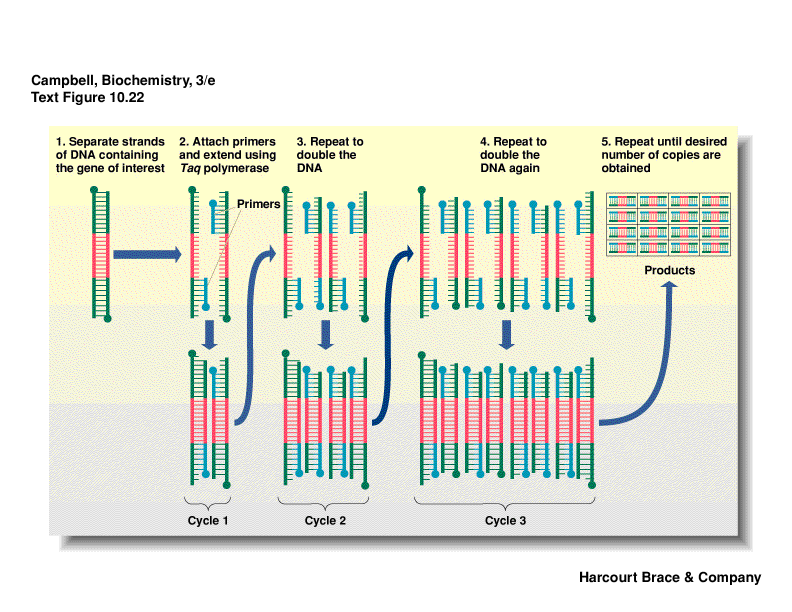
PCR--polymerase chain reaction**
-
easy way to amplify specific regions of DNA
|
 |
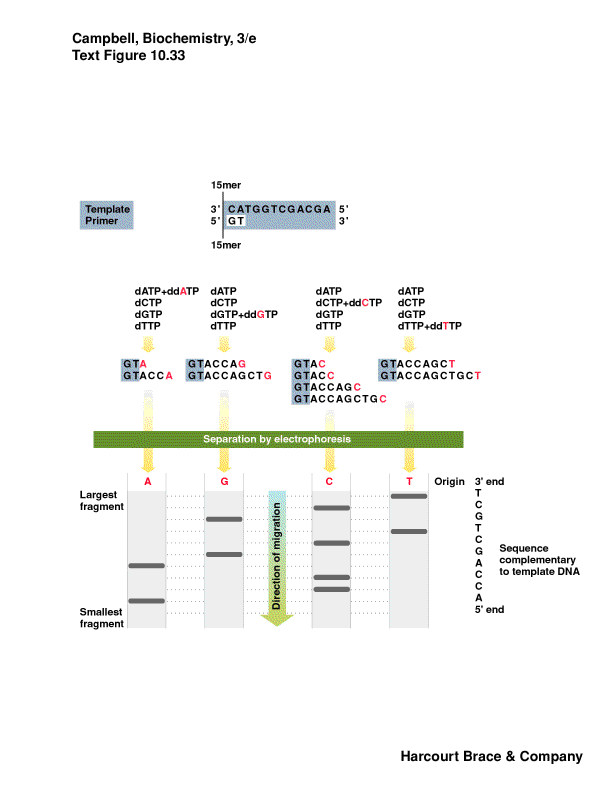
DNA sequencing**
-
determine the precise order of the bases
-
uses DNA polymerase, DNA primer (synthetic oligonucleotide of defined sequence),
radioactive or fluorescently labeled dXTPs
-
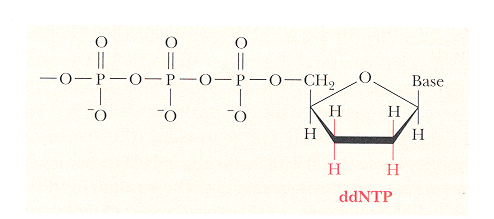
low levels of dideoxy nucleotides to randomly stop elongation of DNA--once
dideoxy is incorporated, no OH for chain elongation
-
represent every possible position
-
separate on a gel to see relative positions and read by "ladder"
sequencing method
|
 |
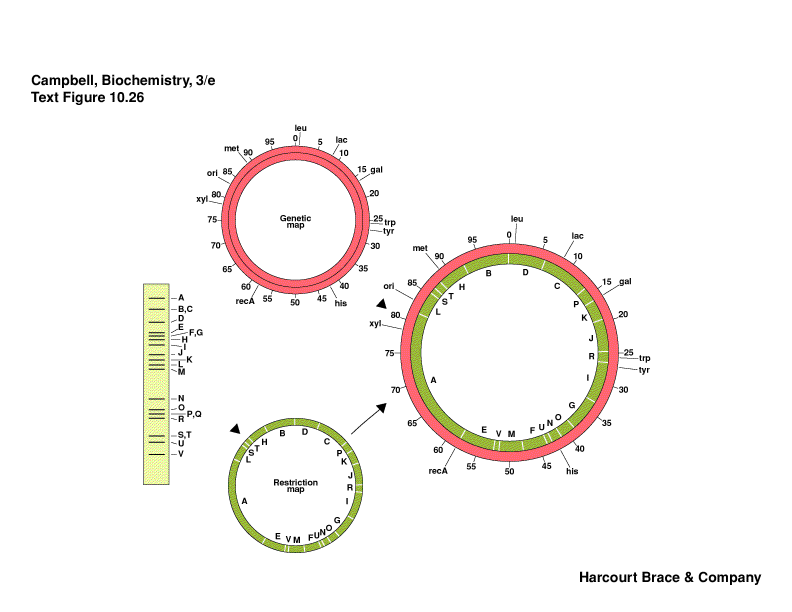
Gene Mapping
-
where is a gene on a chromosome
-
bacterial genes were mapped on how long it took for the gene to be transferred
into another bacteria during conjugation
-
bacteria can exchange genetic information
-
problem with antibiotic resistance and toxin genes
-
compare the genetic map wit the physical map made from restriction fragments
|
 |
Mapping of eukaryotic chromosomes more difficult
-
chromosome walking--end to end matching of DNA fragments
-
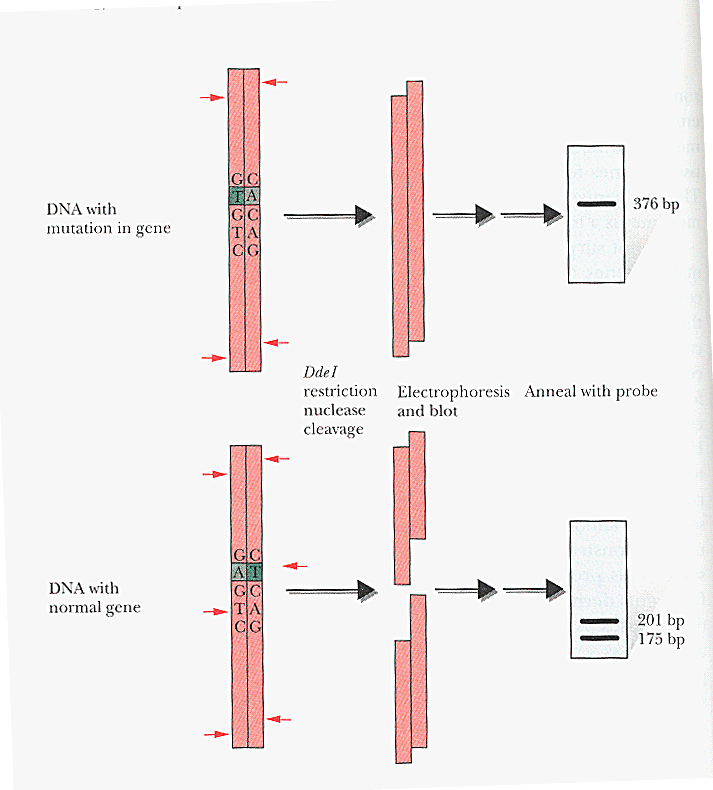
RFLPs--2 chromosomes--DNA in may vary in RE site from one chromosome to
the other
-
lose a site or gain a site due to mutation in a gene
-
different lengths of "filler" DNA
-
distinguish individuals by cutting DNA with RE and comparing patterns--forensics
-
this can be done with genomic DNA and detect with a probe
-
use PCR to amplify a specific region--compare sizes of product
|
 |
|
|














