Nucleosides
/ Nucleotides / Nucleic Acids
Goals for this review unit:
1.
Recognize the common building blocks of nucleic acids – name / 1-letter
abbrev.
2.
Nomenclature for nucleosides and nucleotides (structure of ATP)
3.
Primary structure of RNA and DNA
4.
Conformations in DNAs and RNAs
5.
Characteristics of
B-DNA, A-DNA and Z-DNA
6. Denaturation of DNA
7. Features of RNA /
Functions of RNA
Role of Nucleic Acids
(figure)
Hydrolysis of Nucleic Acids (RNA
--> N base + 5 C ribose sugar + Pi)
- Nucleotides as : building blocks /
cosubstrate /
sec. messenger / transducer
- Nucleoside = N base +
ribose sugar
-
Nucleotide = N base +
ribose sugar +
phosphate
N base ( pyrimidine /
purine )
ribonucleotides (RNAs ;
ribose sugar)
deoxyribonucleotides (DNA ; deoxyribose sugar)
N Bases: Pyrimidines and
Purines (substituted) /
tautomeric forms (know structures)
-
Purines : A = Adenine ;
G =
Guanine; 2 rings (found in RNA and DNA)
-
Pyrimidines : U = uracil ; T = Thymine ;
C = Cytosine
one ring-- C found in RNA and DNA; U in RNA; T in DNA
Nucleosides (deoxynucleosides)
-
N base +
ribose sugar ( deoxyribose
sugar )
- OH group of ribose esterified with phosphate
-
b-N-glycosidic bonds (C1’
to N1 of pyrimidine or N9 of purine)
-
Nomenclature :
Adenosine /
deoxyadenosine (dA)
Guanosine / Uridine / Thymidine /
Cytidine
Nucleotides (deoxynucleotides)
-
Nucleotides = nucleoside - 5’ phosphates
-
monomers connected by covalent bond to form polymers DNA and RNA
-
purine or pyrimidine ring
-
nitrogen containing, weak base--hence the term "bases" when referring to
the nucleotides
-
nitrogens can also ionize at different pHs
-
free bases are rarely found in nature--extremely insoluble
-
double bonds absorb UV--used to quantitatively assay for nucleotides/nucleic acids
Diphosphates and triphosphates --ATP is given as an example
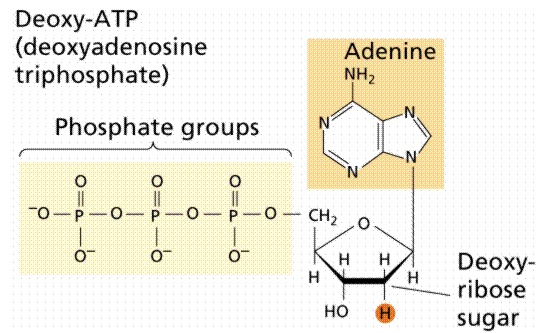 |
-
the phosphate groups are given the designation a (alpha
closest to ribose), b (beta, middle),
g
(gamma, farthest from ribose)
-
di- and tri-phosphates always complexed with Mg++
-
di- and tri-phosphates are used in many metabolic reactions
-
di- and tri-phosphates are synthesized from the monophosphates by kinases
using ATP as the source of phosphate
|
|
|
ATP (triphosphate)
|
|
|
Nucleic
Acids:
-
polymers: Two types:
DNA = deoxyribonucleic acid /
RNA
= ribonucleic acid
-
monomer units connected by covalent (phosphodiester) bond
-
oligonucleotides--short pieces of DNA or RNA
Primary
Structure of NAs
-
directionality--phosphodiester linkage is 3' to 5' with
a 5' phosphate and a 3' OH
-
nucleotide sequence = primary structure
-
genetic information is stored in the primary sequence
-
nucleic acid sequence is written 5' to 3' - 5'
pAGCTAAGGCCTTTACTAG OH 3'
Deoxyribonucleic acids (DNA)
-
backbone of alternating units of 2-deoxy-ribose and phosphate in
which the 3’-OH of one deoxy-ribose is joined by a phosphodiester bond
to the 5’-OH of another deoxy-ribose unit
-
Primary Structure: the sequence of bases along the pentose-phosphodiester
backbone of a DNA molecule (or an RNA molecule)
-
5’ end to the 3’ end--directionality
|
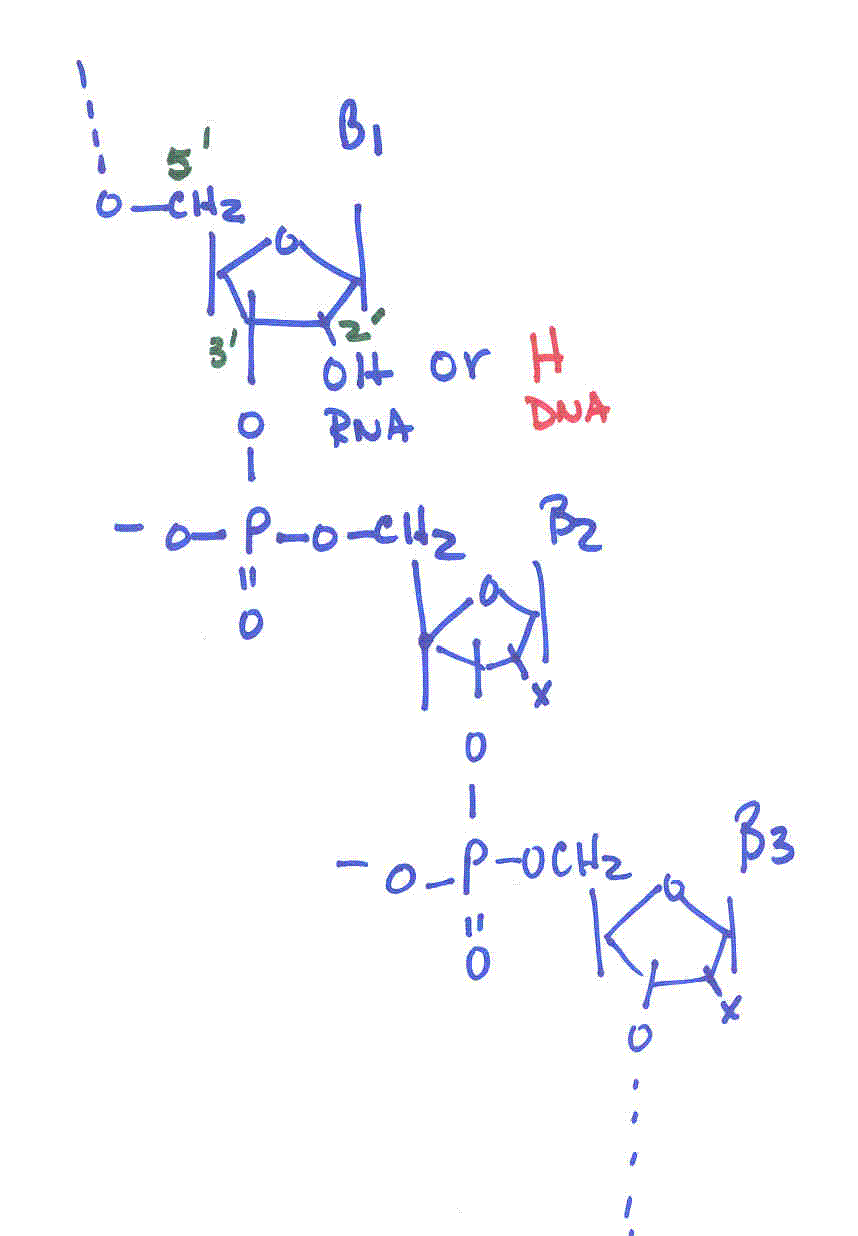 |
Secondary structure: the ordered arrangement of nucleic acid
strands
-
the double helix model of DNA 2° structure was proposed by James Watson
and Francis Crick in 1953
-
Double helix: 2° structure of DNA molecules in which two antiparallel
(5'-->3'/ 3'-->5') or complimentary polynuceotide strands are coiled in
a right-handed manner; C2'-endo sugars
-
stabilizing the double helix is base pairing between T-A and between
C-G
by hydrogen bonds and base stacking
-
Which is the stronger base pair? Why?
-
phosphate backbone position minimizes repulsion of negative charges
-
There are other nucleotide pairings: Wobble, Hoogsteen, reverse
Hoogsteen, G-quartet, etc.
|
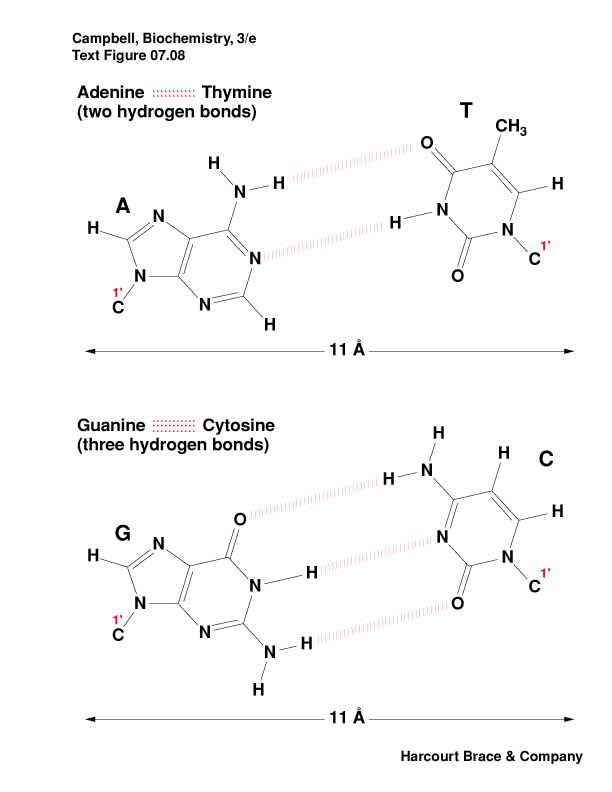 |
Helical
DNA
The
Double Helix--1953 - B-DNA
- Erwin Chargaff : %G=%C and
%A=%T
-
X-ray diffraction of DNA by Rosiland Franklin
-
Watson and Crick recognized that this was a type of helical structure
spacings
were 1/10 of pattern repeat, 10 residues/turn of the helix
densities
suggested that there were two DNA strands/molecule
hydrogen
bonding between the bases would stabilize the structure
base
pairing had to be only one way: G=C and A=T
means
the two strands are always complementary
phosphate/ribose
backbone on the outside in contact with solvent
bases
inside helix strands
have to run in opposite direction 5' to 3' and 3' to 5'
bases
may be approached from either minor or major groove
Conformations
of Nucleic Acids
-
Conformations of nucleosides - syn / anti
for relative positions
of N base vs. ribose sugar ring (figure)
- Sugar pucker: ribose sugar rings typically
are five membered rings with 4 of the 5 atoms nearly planar. The
atom displaced can be endo (within - same side as C5' or N base) or exo (out of
- opposite side)-
C3' endo / C2' endo
/ C3' exo most common (see DNA table below)
- There are preferred torsion angles for nucleic acids like for proteins - just
more of them.
Nucleic Acid Gallery--
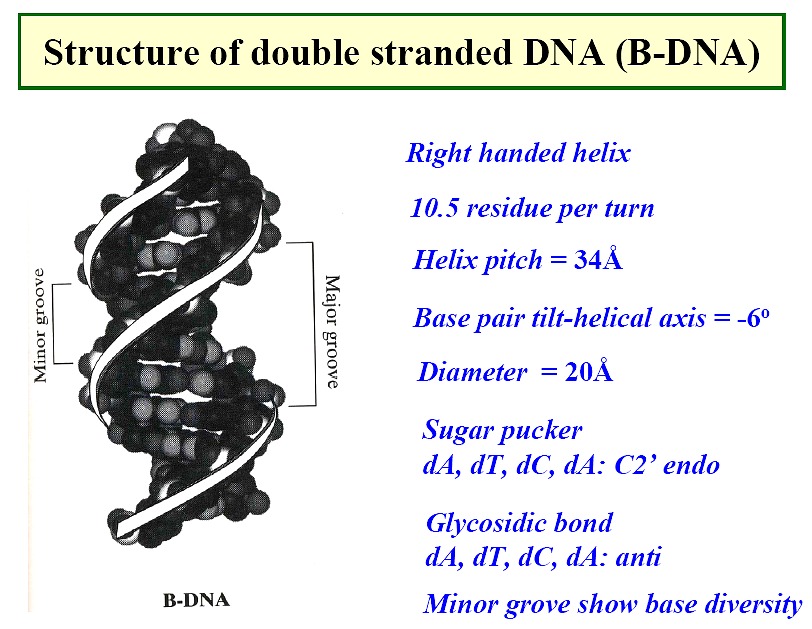 |
 |
 |
B form DNA
-
the predominant form in dilute aqueous solution
-
a right-handed helix
-
34Å per 10 base pairs; 20Å diameter
-
minor groove of 6Å and major groove of 12Å
-
sugar - anti; pucker - C2'-endo with PO4-PO4
distance of 7.0Å
-
positively charged ions, Mg++ or Na+ and positively charge proteins (histones)
bind through electrostatic interactions
|
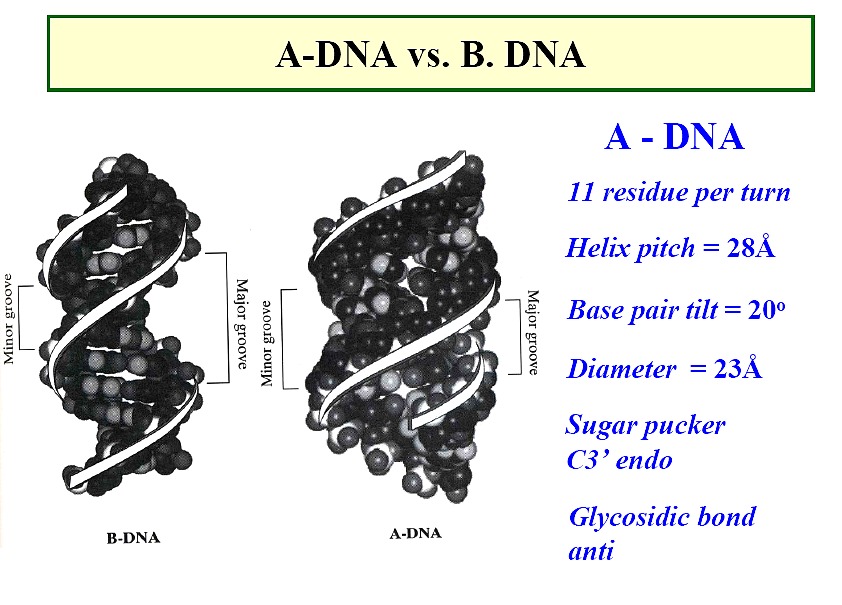
A form DNA
-
depends on conditions, such as ions
-
a right-handed helix, but thicker than B-DNA, 26Å diameter
-
29Å per 11 base pairs
-
tilt ~20o , rise ~2.6Å
-
sugar - anti; pucker - C3'-endo with PO4-PO4
distance of 5.9Å
|
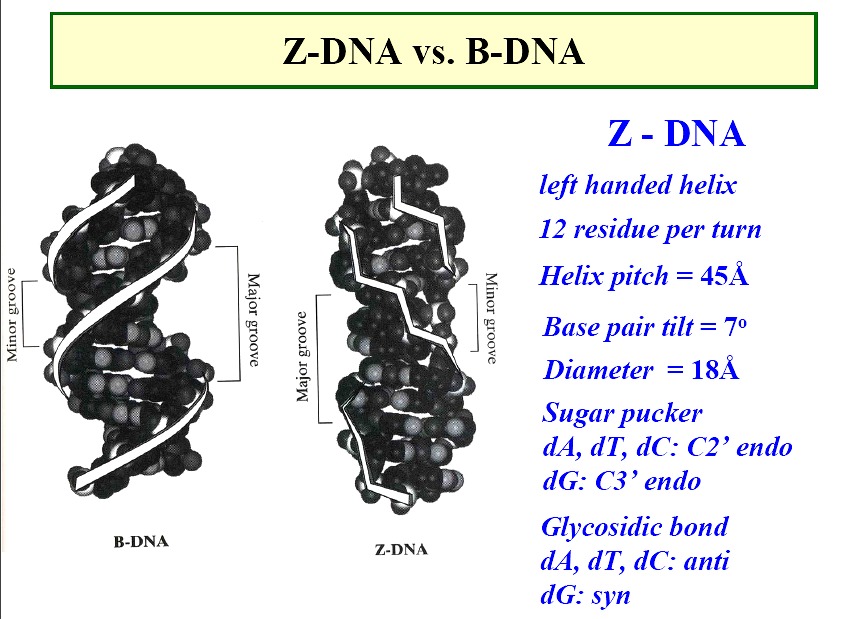
Z form DNA
-
a left-handed double helix; "zig-zag"
-
forms in nature--function unknown, maybe gene expression regulation
-
diameter ~18Å
-
45Å per 12 base pairs
-
sugars
- - pyrimidines - anti; pucker -C
C2'-endo
- - purines - syn; purines -G
C3'-endo
|
Stabilization (destabilization) of DNA Structure
-
Hydrogen Bonding - more important for "specificity" than overall
stability
-
Electrostatic Repulsion - anionic phosphate backbone
-
Stacking - Induced dipole interactions between aromatic bases
-
Solvent interactions - Observe a "spline" of ordered water molecules
in grooves.
Denatuted DNA:
Heat denaturation of DNA is called
"melting," this
usually occurs in a "cooperative" manner.
Absorption spectroscopy can be used to monitor the "helicity" of
nucleic acids. The purine and
pyrimidine bases exhibit very strong p-p*
transitions around 260 nm. The degenerate interactions
between bases causes splitting and redistribution of intensity, the
nondegenerate interactions can lead to
a loss of intensity. E. coli DNA absorption is only about 60% of that
predicted from the weighted average
spectrum based on its composition, this loss of intensity is called hypochromism.
Since the absorpance
goes up as DNA "unwinds", it can be used to monitor the unstacking of
DNA. Renaturation on cooling is
incomplete since the DNA will assume many partially folded states (see pp
402-408 of text.).
DNA denaturation
-
separate strands of DNA--melting
-
must break H bonds and disrupt base stacking
-
heat or urea
-
remember nucleotides absorb in UV range due to ring structure
-
base stacking reduces amount of light absorbed
-
pull apart strands, increase amount of light absorbed
-
following melting of DNA by UV
-
denaturation important in replication and manipulation of DNA in laboratory
|
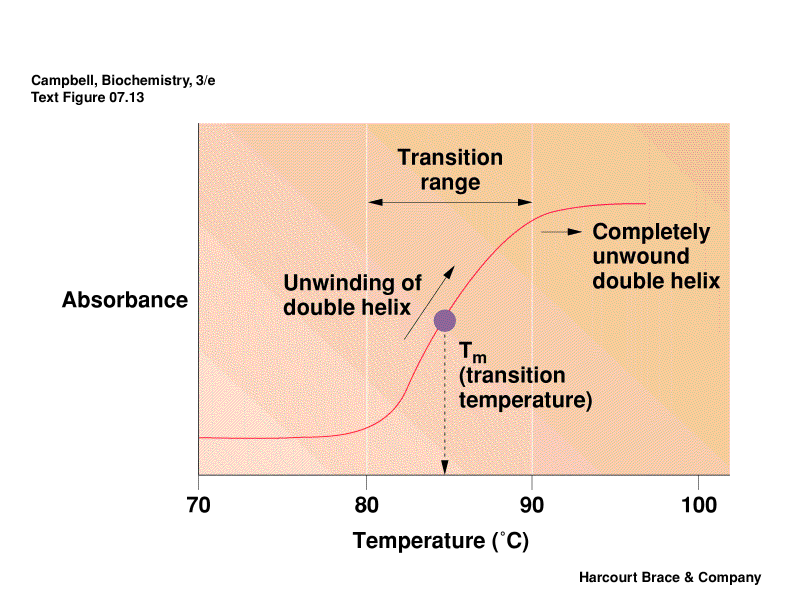 |
Tm
Values: Define Tm as the temperature at which half the melting has
taken place. The degree of
hypochromism depends on its sequence, ionic environment, pH, etc. Tm
values usually rise with increasing
length of n.a., G/C content, and increasing salt concentration due to less repulsion by
anionic phosphate backbone.
At the Tm, the concentrations of NAfolded = NAunfolded,
so thermodynamic data can be obtained.
Sequence
dependence of Helical Stability:
It can be shown that G=C base pairs are more stable that A=T base pairs,
etc. In fact, the correlation between
calculated and observed Tm values for defined
sequences is very good, thus one can make good estimates at
predicting helix stability. This has
practical implications in several ways: 1) estimate temperature for probe
stability,
2) means that one can use computers to estimate the free
energies associated with various "folds" of the nucleic
acid and have a better chance of predicting secondary
structure (see textbooks for structures of tRNAs and rRNAs),
and 3) we can design molecules of n.a. to bind and render other n.a.'s inactive
(antisense drugs).
RNAs
-
Classical picture: m-RNA (Figure),
t-RNA (Figure), r-RNA (ribosome)
Ribozymes - e.g. tetrahymena ribozyme (Figure1,
Figure2, Figure3,
Figure4)
Use of FRET to study RNAs (Figure1, Figure2,
Figure3)
RNA world view (Figure)






