Protein Expression, Purification and Analysis
To
study proteins and their functions, one must first Produce,
Extract, and Purify the protein.
Produce - tissue
rich in protein / over-expression using cultured cells (see below)
Extract - cell
disruption followed by centrifugation
Purification -
take advantages of differences in solubility, charge, size, and specificity.
(An assay
is needed to monitor the progress of the purification process.)
********************************
Purification
based on solubility and charge:
Protein purification used to start with a source (organ
tissue, plant source, bacteria line) known to be rich in the protein of
interest. Often this would involve
obtaining many pounds of organ tissue directly from a slaughter house or growing
large (30 or 60L) batches of bacterial culture in order to isolate sufficient
material for further studies.
Typical protocol for isolation of a mammalian protein (after
procedure for chicken heart LDH – H4, Nathan
Kaplan et al., JBC, 239: 1753-1761 (1964):
***********************
Modern Methods: Today, the majority of proteins being studied in the laboratory
take advantage of more modern tools of biotechnology to produce
large quantities of proteins needed for study.
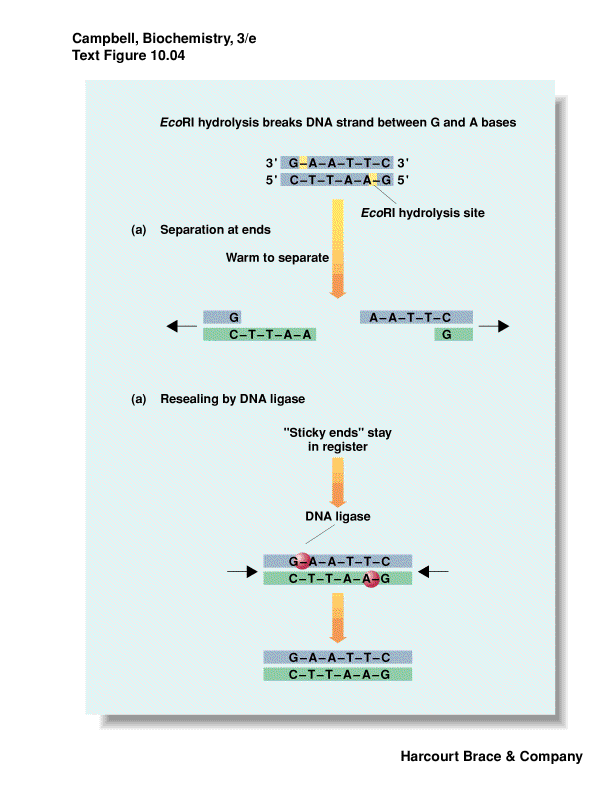
Restriction enzyme--molecular scissors
-
endonucleases--does not require an end (exonucleases)
-
>100 restriction enzymes known
-
names come from organism:
-
recognize a specific palindromic DNA sequence and cut the DNA
-
palindrome is the same forwards/backwards
-
some leave 3' overhang; 5' overhang or blunt ends
-
overhangs leave--"sticky ends"--even though DNA is cut, can have base-pairing
-
move DNA from one organism to another - "recombinant DNA"
-
put DNA together with DNA ligase
-
use synthetic DNA of desired sequence to "paste" on restriction site if
nature did not provide one
|
-
methylation protects DNA from restriction enzymes
-
mechanism for bacteria to protect itself from invading phage or other bacterial
DNA
 |
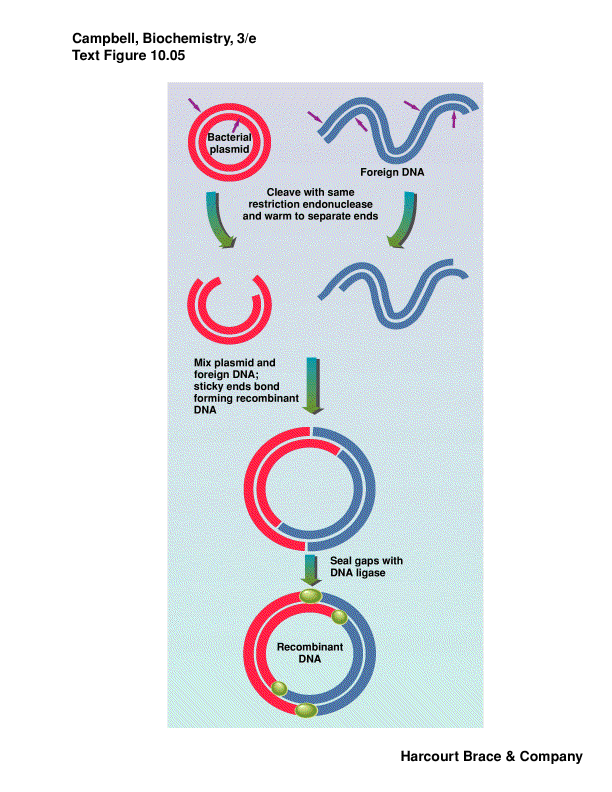
Plamids are cloning vectors
-
plasmids are closed circular DNA, with origin of replication--replicated
within bacteria to many copies
-
carries a resistance gene--ampicillin, tetracyclin, kanamycin
-
take DNA from one organism, cut with RE, isolate fragment desired from
a gel
-
cut a plasmid or phage DNA with same RE
-
put these two DNA fragments together via sticky ends, ligate them closed
-
we have recombinant DNA
-
this is transferred into bacterial cells by electroporation or chemical
competence
-
plate on media with antibiotic to kill bacteria that did not take up a
plasmid--no proof that your foreign DNA is there, only that the plasmid
is there
-
individual colonies contain a single plasmid
|
 |
-
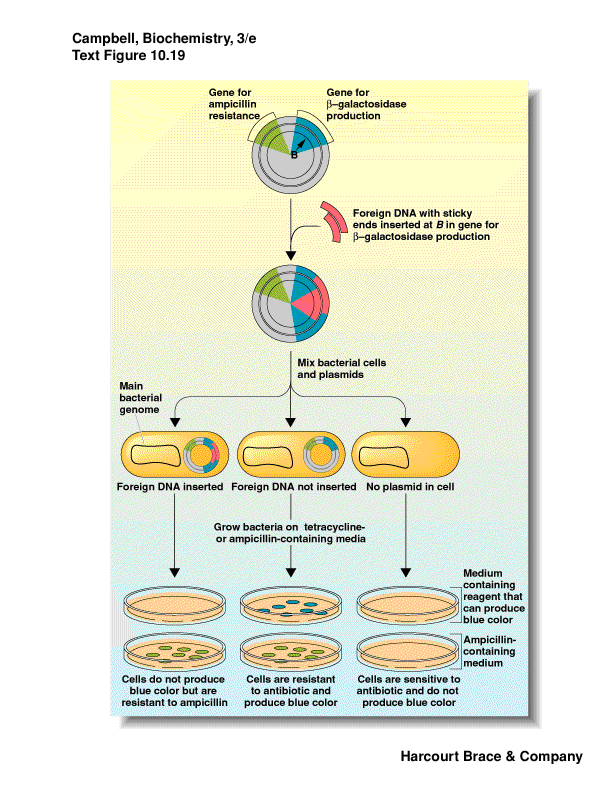
How do you know your foreign DNA was inserted?
-
one method: interrupt a gene that is a reporter - b-galactosidase (lacZ)
-
use a substrate for b-galactosidase that when
cleaved give a colored compound
-
do this on antibiotic media to select for plasmid
-
induce the gene with a lactose-analog
-
if the gene is intact get blue color--no foreign insert, just plasmid
-
if the gene has an insert (foreign DNA) then the reading frame is thrown
off and no b-galactosidase is produced--no color
|
 |
Purification -
take advantages of differences in solubility,
charge, size and
specificity.
1. Solubility
2.
Charge: column
chromatography
- Separation by charge (size
or
affinity)
-
a matrix is in a cylindrical holder
-
buffer flows through the matrix
-
fractions are collected
-
separation of biomolecules
Charge:
Ion-exchange chromatography
-
proteins have charges due to amino acid side groups
-
bind to charged column matrix depending on their charge at a particular
pH
-
anionic matrix--negatively charged (cation exchanger): CMC (carboxymethyl
cellulose), phosphocellulose, heparin sepharose, S-sepharose
-
cationic matrix --positively charged (anion exchanger): DEAE-sepharose, Q-sepharose
-
elute bound proteins from column based on charge and displacement by salt
or pH
High Performance Liquid Chromatography
(HPLC)
-
gravity flow very slow--depends on size and amount of liquid at the top
-
HPLC used high pressure to force liquid through
-
special matrixes and columns
-
fast and sometimes better resolution
3. Size -
i) Dialysis
(figure)
-
separates on the basis of size, not charge
-
porous beads--think of golf balls
-
small molecules go into the holes and get trapped temporarily (Figure)
-
large molecules are too large to enter the holes and pass on by
-
exclusion size--depends on the size of the holes
-
how long the molecules get trapped determines elution order
-
large out first > medium > small out last (Figure)
-
choose the size of matrix for the separation needed
-
Terms: Vtot, Vo, Vpoly, Ve, Kav = (Ve - Vo)/(Vt - Vo)
-
Plot Kav vs. log MW for known standards and unknown to estimate MW
4. Specificity:
Affinity Chromatography - use of "tagged"
proteins to create affinity site - sep. by specificity
-
column matrix has a ligand that specifically binds a protein
-
specialty affinity columns for binding recombinant proteins with certain
"tags"
-
6xHis added at N or C terminus--binds Ni++ column
-
His tag (Figure 1) (Figure
2) (Figure 3) (Figure 4)
-
other types of "tags"--chitin, glutathione S-transferase (GST).....
|
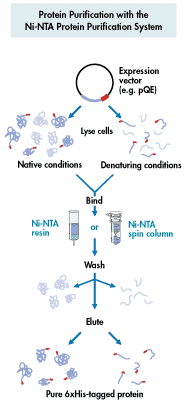
from Qiagen
website |



