This image is by Dr. George Helmkamp, Jr (UKMC).
Protein Structure
Proteins: Biological Function depends on conformation
Unique Primary Structure = Unique 3D Structure
(Covalent bonds)
(Noncovalent Interactions)
Globular
Proteins: water soluble, compact, hydrophobic interior / hydrophilic surface
enzymes, receptors, carriers, hormones, etc. (dynamic
agents)
Fibrous
Proteins: water insoluble, structural roles, extended structure
collagen (tendons, bone), a-keratin
(hair, nails), etc. (~static
agents)
Four Levels of Description of (Native) Protein Structure
Primary Structure: (~60-1000 amino acid residues)
linear
seq. of amino acid residues, covalent bonding including -SS-
(also called "covalent structure") definition
(the primary structure of a biological molecule is the exact specification of its atomic composition and the chemical bonds connecting those atoms (including stereochemistry). In general, polypeptides are unbranched polymers, so their primary structure can often be specified by the sequence of amino acids along their backbone. However, proteins can become cross-linked, most commonly by disulfide bonds, and the primary structure also requires specifying the cross-linking atoms, e.g., specifying the cysteines involved in the protein's disulfide or other covalen bonds.)
Secondary Structure: definition
local conformations of backbone, maintained by hydrogen bonds
Tertiary Structure:
3D structure of a subunit (one polypeptide chain) in its native
state
Quaternary Structure:
Spatial arrangement of subunits in oligomeric proteins
Denaturation:
Partial to complete unfolding of native conformation
Denatured Protein: Protein that has lost its native conformation
This image is by Dr. George Helmkamp, Jr (UKMC).
Primary Structure
Protein Sequencing Strategies
Older Methods
N-terminal
and C-terminal analysis
Trypsin (cleavage after Arg, Lys)
Chymotrypsin (cleavage after aromatic residues, Phe, Trp, Tyr)
Carboxypeptidase - cleaves C-terminal residue
CNBr - chemical cleavage after internal Met residues
Newer Methods
DNA
sequencing (Genetic Code à
Protein Sequence from ORF)
Databases - Nucleic
acid sequences
/ Protein
sequences
- Saccharomyces:
12,068 kb, 5885 genes
- NCBI (Medline, GenBank, Entrez, Blast)
|
This image is by Dr. George Helmkamp, Jr (UKMC) |
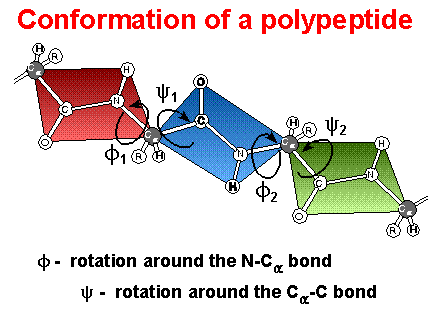
Peptide Bonds / Peptide Conformation
Peptide bond: ~ double bond character; planar group, trans conformation (cis ? proline)
Peptide
conformations:
Phi / Psi ( f
/ y
) angles; Ramachandran
plots - “allowed” angles
- Definition of Phi / Psi ( f / y ) torsion or dihedral angles. Note: The definitions below come from IUPAC - IUB rules for biochemical nomenclature (Biochemistry 9:3471 (1970)). Some times you will find torsion angles defined differently but the values tabluated below will be identical.
f : torsion angle for Ni-Cia
f is considered (+) when the system is viewed down the central bond (Ni-Cia) and the bond in front (Ci-1' - Ni) requires a rotation to the right (clockwise) in order to that it eclipse the bond to the rear atom (Ci').
y : torsion angle for Cia - Ci'
y is considered (+) when the system is viewed down the central bond (Cia - Ci') and the bond in front (Ni-Cia) requires a rotation to the right (clockwise) in order to that it eclipse the bond to the rear atom (Ni+1').
f
= y
= 180o (fully extended, planar conformation)
f
= -57o ;
y
= -47o (right handed a
helix)
f
= -139o ;
y
= +135o (antiparallel b
sheet)
f
= -119o ;
y
= +113o (parallel b
sheet)
Secondary
Structure of Proteins (local folding / periodic structure of protein
backbones)
- a-helix
: (Linus Pauling & Robert Corey - 1951;
Diff. a-keratin
pitch = 0.54 nm; rise 0.15 nm; 3.6 residues / turn
helix formers:
Met, Glu;
helix breakers: Pro, Gly
helical wheels / amphipathic helices:
(n, n + 4, n + 7, etc.)
- bab
unit / helix-loop-helix
:
- Hairpin loop:
¯ (antiparallel);
Cross-overs
- Greek key motif : ¯
¯
- b
barrel
Tertiary Structure - 3D structure of polypeptide chain
Forces
Stablizing Tertiary Structure
- Hydrogen bonding (backbone
and sidechain)
- Hydrophobic interactions
- "Ion pairs" - Electrostatic interactions
- Disulfide bonds
Tertiary Structure of Globular Proteins
- hydrophobic AA generally on the inside
- hydrophilic AA generally on the outside in contact with water
Domains
: Conbination of motifs - 25 to
300 a.a. / function
- b
sandwich /
b
barrel / a/b
barrel /
helical bundle
Functional
units
- Some Unusual Folding patterns of proteins are unique to their function
- left-handed parallel beta helix / horseshoe crab
- Nucleotide binding domain babab
/ dinucl. binding domain = Rossmann Fold
- Zn finger / Leucine zipper
Some Examples of Protein Structures (pink = helices; yellow = sheets)
|
Bovine Pancreatic Trypsin Inhibitor-
|
Porin-beta barrel |
Prealbumin-Greek key |
Triose Phosphate Isomerase (TIM) bab barrel |
Quaternary
Structure - Arrangements of subunits in “oligomers”
a4 ; a12
; (ab)2
; (ab)6
Many oligomeric proteins are allosteric (Mb vs. Hb - next unit)
|
4-oxalocrotonate tautomerase (Monomer) |
4-oxalocrotonate tautomerase (Dimer) |
4-oxalocrotonate tautomerase (Hexamer) |
More sites with protein structure tutorials:
CMU Protein
Architecture Site
Protein
Architecture
Note: Some of the figures used in these class notes are copyrighted material from the text and are not to be used for any other purpose than to support this course.